Code
# load dependencies
library(ggplot2)
library(tidyverse)
library(purrr)
library(cowplot)
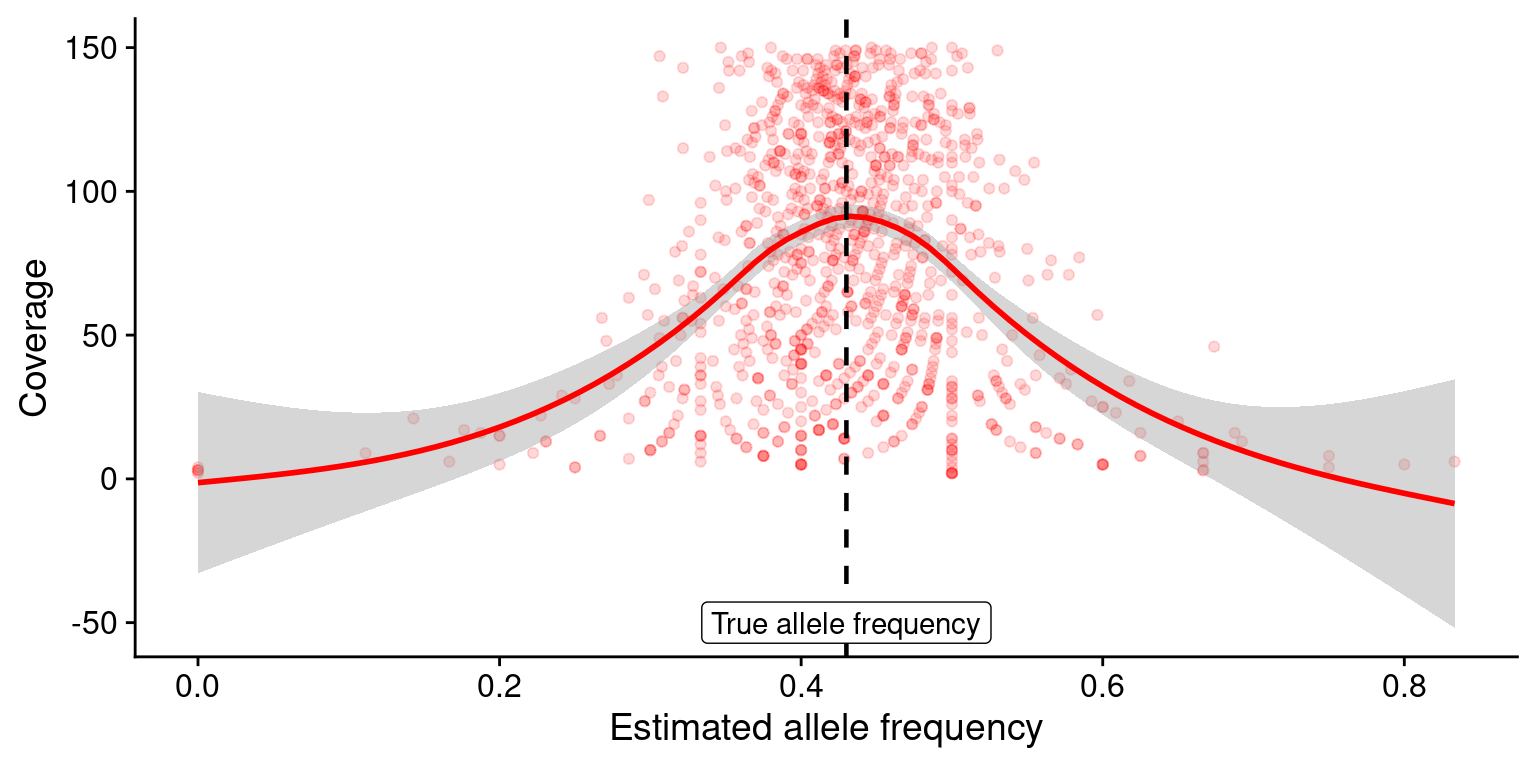
set.seed(172452)Simulate a pairwise comparison between alleles at a single location with coverage error. To do this, we use a random binomial distribution that takes in coverage and true allele frequency and outputs the estimated allele frequency.
[1] 0.45Function that calculates estimated allele frequency when it is given coverage and true allele frequency:
Now, we can scale our code and plot the results:
# run this n times, scale up
n <- 1000
tru <- 0.43
cvg <- sample(2:150,n,replace = TRUE)
af <- tibble(
"cvg" = cvg,
"trueAF" = rep(tru,times=n)
)
af$estAF <- af %>% pmap(estAF) %>% unlist()
# plot estimated allele frequency by coverage
plt <- af %>% ggplot(aes(x = estAF, y = cvg)) +
geom_point(alpha = 0.15,colour="red") +
geom_smooth(method = "gam", colour = "red",se = FALSE) +
geom_vline(xintercept=tru, size = 0.8,linetype = "dashed") +
labs(x = "Estimated allele frequency", y = "Coverage") +
annotate("label",x = tru, y = -50, label = "True allele frequency") +
theme_cowplot()
plt